Who produces methylmercury in the global ocean?
The UNEP Minamata Convention was ratified in August 2017 and aims to protect human health from mercury exposure by reducing anthropogenic, inorganic, mercury emissions. The most toxic and biomagnifying mercury species, methylmercury, is not emitted from anthropogenic or natural sources, but produced in the ocean from inorganic mercury.
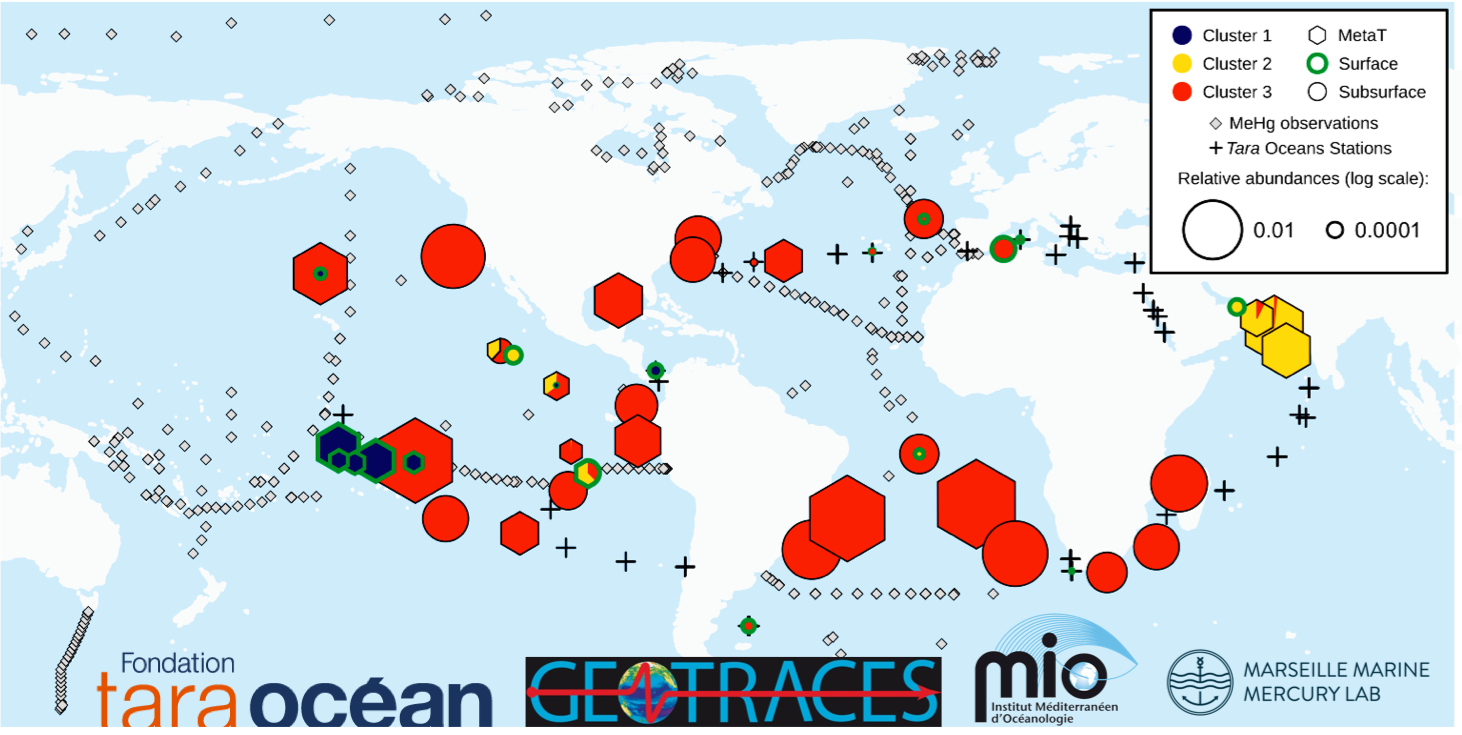
Marine mercury methylation has been confirmed for most ocean basins independent of seawater oxygen levels. Yet, only anaerobic organisms are known to possess the genes required for mercury methylation. This makes the origin of marine methylmercury somewhat of a paradox and the question of who or what methylates mercury in the ocean remained unresolved. Solving this question is of prime importance to better understand the links between mercury emissions and the propagation of mercury along the marine food web, that puts ecosystem and human health at risk.
A group of young researchers of the Mediterranean Institute of Oceanography (MIO) triangulated this outstanding question with their combined expertise in bioinformatics, microbiology and marine biogeochemistry. Based on an intensive metagenome sample set from the tara oceans project they find mercury methylation genes corresponding to taxonomic relatives of known mercury methylators from Deltaproteobacteria, Firmicutes and Chloroflexi phyla in every covered ocean basin. The well-known anaerobic sulfate-reducing bacteria, usually considered to be the dominant methylmercury producers, represent only a minor fraction in seawater. The results identify Nitrospina, a microaerophilic nitrite-oxidizer, as the predominant and ubiquitous methylmercury producer in the oxic subsurface waters of the global ocean (Pacific, Atlantic, Indian and Southern oceans).
The findings resolve the long-lasting paradox of abundant marine methylmercury production in the absence of the known anaerobic methylmercury producers. It is surprising to find an aerobic bacterium responsible for a supposedly anaerobic process, such as mercury methylation. The results are a significant contribution to the understanding of the global mercury cycle and have important implications for the potential impacts of climate change on marine biodiversity and as such on marine methylmercury production.
Reference:
Villar, E.; Cabrol, L.; Heimbürger‐Boavida, L. Widespread Microbial Mercury Methylation Genes in the Global Ocean. Environ. Microbiol. Rep. 2020, 1758-2229.12829. DOI: https://doi.org/10.1111/1758-2229.12829
